 Author
Author  Correspondence author
Correspondence author
Animal Molecular Breeding, 2024, Vol. 14, No. 4
Received: 02 May, 2024 Accepted: 20 Jun., 2024 Published: 05 Jul., 2024
This study explores the foundational principles of quantitative genetics, including Mendelian inheritance, polygenic traits, heritability, and genetic variance, and examines their practical applications in breeding programs. Key tools and methodologies, such as quantitative trait loci (QTL) mapping, genome-wide association studies (GWAS), and selection indices, are discussed in detail. Applications in enhancing productivity traits, improving health, and optimizing reproductive efficiency are highlighted, with a case study on dairy cattle breeding illustrating the economic and practical impacts of heritability and breeding value estimates. Emerging trends, including the integration of genomics, artificial intelligence, and CRISPR-based precision breeding, are addressed alongside challenges such as environmental interactions, ethical considerations, and the cost of advanced tools. The paper concludes by emphasizing the critical role of quantitative genetics in sustainable livestock breeding and advocating for continued research to address future challenges and opportunities.
1 Introduction
Quantitative genetics is a branch of genetics that deals with the inheritance of traits that are determined by a large number of genes, each contributing a small effect, and are often influenced by environmental factors. These traits, known as quantitative traits, exhibit continuous variation and are typically measured on a numerical scale, such as body weight, milk yield, or egg production in livestock (Schmid and Bennewitz, 2017; Silva et al., 2017; Núñez-Torres and Almeida-Secaira, 2022). The scope of quantitative genetics extends to understanding the genetic architecture of these traits, predicting breeding values, and improving selection methods to enhance desirable traits in livestock populations (fHill, 2010; Rukundo et al., 2018).
Quantitative genetics plays a crucial role in livestock breeding by providing the theoretical foundation and practical tools necessary for the genetic improvement of economically important traits. The application of quantitative genetics in livestock breeding involves the use of statistical models to predict breeding values, which are essential for selecting animals with superior genetic potential (Haley, 2002; Núñez-Torres and Almeida-Secaira, 2022). This field has significantly contributed to the efficiency of livestock production by enabling breeders to make informed decisions based on genetic data, thereby optimizing the genetic quality of livestock (Gautier, 2001; Jenko et al., 2015). Moreover, advancements in genome-wide association studies (GWAS) and genomic selection have further enhanced the ability to map and exploit quantitative trait loci (QTL) for improved breeding outcomes (Schmid and Bennewitz, 2017).
This study attempts to elucidate the principles of quantitative genetics and their application in livestock breeding, discuss the importance of quantitative genetics in enhancing livestock production, and provide an overview of the latest advancements and methodologies in the field. By examining the integration of quantitative genetics with modern genomic tools, this study aims to provide insights into the future directions of livestock breeding programs and the potential for continued genetic improvement.
2 Theoretical Foundations of Quantitative Genetics
2.1 Mendelian principles underlying quantitative traits
Quantitative traits in livestock are influenced by multiple genes, each contributing a small effect, which aligns with Mendelian principles of inheritance. These traits, such as body weight gain and milk production, exhibit continuous variability and are typically distributed normally due to the additive effects of numerous genes. The action of additive genes results in a phenotypic distribution that is intermediate between the parental populations, while multiplicative gene actions can create geometric series in trait expression (Núñez-Torres and Almeida-Secaira, 2022). Understanding these principles is crucial for developing effective breeding strategies that enhance economically important traits in livestock.
2.2 Polygenic inheritance and complex trait analysis
Polygenic inheritance refers to the genetic architecture where multiple genes contribute to the expression of a single trait. In livestock, most economically significant traits are complex and influenced by numerous genetic and environmental factors. Genome-wide association studies (GWAS) have become a pivotal tool in identifying quantitative trait loci (QTL) that contribute to genetic variance in these traits. The use of dense SNP panels allows for the mapping of genes and mutations that affect these complex traits, providing insights into their genetic underpinnings and facilitating more precise selection in breeding programs (Schmid and Bennewitz, 2017; Singh et al., 2019; Bijma, 2021).
2.3 Concepts of heritability and genetic variance
Heritability is a key concept in quantitative genetics, representing the proportion of phenotypic variance that can be attributed to genetic variance within a population. It is a critical factor in determining the potential for genetic improvement through selection. Genetic variance encompasses both additive and non-additive genetic effects, with additive variance being particularly important for selection as it directly contributes to the response to selection. Understanding the genetic variance and heritability of traits allows breeders to predict the outcomes of selection and optimize breeding strategies to enhance desirable traits in livestock populations (Figure 1) (Viana et al., 2016; Núñez-Torres and Almeida-Secaira, 2022; Kandel et al., 2023).
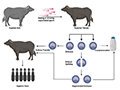 Figure 1 Schematic presentation of multiple ovulation embryo transfer in buffalo (Adopted from Kandel et al., 2023) |
In summary, the theoretical foundations of quantitative genetics in livestock breeding involve understanding Mendelian principles, polygenic inheritance, and the concepts of heritability and genetic variance. These principles guide the development of breeding strategies aimed at improving economically important traits through informed selection and genetic analysis.
3 Tools and Methods in Quantitative Genetics
3.1 Quantitative trait loci (QTL) mapping
Quantitative Trait Loci (QTL) mapping is a fundamental tool in quantitative genetics, used to identify genomic regions associated with the expression of quantitative traits. These loci may contain one or more genes that contribute to the trait's variance. QTL mapping typically involves analyzing biparental populations to detect associations between marker genotypes and trait phenotypes. Various methods, such as regression analysis, maximum likelihood estimation, and Bayesian models, are employed to identify QTLs. Single QTL mapping methods focus on detecting one QTL at a time, while multiple QTL mapping combines multiple regression analysis with interval mapping to include all significant QTLs in the genetic model. This approach can be extended to handle data from multiple cross populations and for joint analysis of multiple traits (Singh and Singh, 2015). Fine mapping of QTL regions is often achieved using homozygous lines derived from near-isogenic lines (NILs) and intercross recombinant inbred lines. Additionally, QTL meta-analysis integrates results from different studies to refine the number of QTLs affecting a trait and reduce confidence intervals (Halladakeri et al., 2023).
3.2 Genome-wide association studies (GWAS)
Genome-Wide Association Studies (GWAS) have become a preferred method for mapping quantitative traits in livestock due to the availability of dense SNP panels. GWAS involves scanning the genome to identify SNPs associated with traits of interest. Different statistical models, including single-marker and Bayesian multi-marker models, are used in GWAS to account for nonadditive genetic effects and genotype-by-environment interactions (Schmid and Bennewitz, 2017). Multibreed GWAS can enhance mapping precision and power by leveraging conserved linkage disequilibrium across breeds, as demonstrated in dairy cattle studies. This approach can detect more QTLs compared to within-breed GWAS, especially when multiple populations are combined (Berg et al., 2016). GWAS results can be further refined through conditional analyses and fine mapping to identify significant SNPs and candidate genes (Li et al., 2020).
3.3 Selection indices and breeding values
Selection indices and breeding values are critical tools in livestock breeding, enabling the selection of individuals with desirable genetic traits. These indices combine information from multiple traits to provide a comprehensive measure of an individual's genetic potential. Breeding values are estimated using statistical models that incorporate phenotypic and genotypic data, allowing breeders to predict the genetic merit of animals for specific traits. The integration of genomic data into selection indices has improved the accuracy of breeding value predictions, particularly for complex traits like fertility in dairy cattle (Ma et al., 2019). Genomic selection has stabilized and even reversed unfavorable trends in traits such as dairy fertility, demonstrating its effectiveness in modern breeding programs.
In summary, QTL mapping, GWAS, and selection indices are essential tools in quantitative genetics, each contributing uniquely to the understanding and improvement of complex traits in livestock. These methods enable the identification of genetic loci, the prediction of breeding values, and the enhancement of breeding strategies through genomic selection.
4 Applications of Quantitative Genetics in Livestock Breeding
4.1 Enhancing productivity traits: milk, meat, and egg yield
Quantitative genetics plays a crucial role in enhancing productivity traits such as milk, meat, and egg yield in livestock. These traits are typically quantitative, meaning they are influenced by multiple genes and environmental factors, leading to continuous variability in the population (Núñez-Torres and Almeida-Secaira, 2022). Techniques such as genome-wide association studies (GWAS) and marker-assisted selection (MAS) have been employed to identify and select for genes associated with these economically important traits (Khalil and Gonda, 2020). By understanding the genetic basis of these traits, breeders can make informed decisions to improve yield and efficiency in livestock production (Khatib and Gonda, 2015).
4.2 Improving health and disease resistance
Quantitative genetics also contributes significantly to improving health and disease resistance in livestock. Although individual disease traits often have low heritability, the genetic variance for disease prevalence can be substantial due to indirect genetic effects. This implies that selection against infectious diseases can be more effective than previously thought. By integrating quantitative genetics with epidemiology, breeders can develop strategies to enhance disease resistance, thereby reducing the impact of pathogens on livestock (Bijma, 2021). Additionally, genome editing techniques, such as the promotion of alleles by genome editing (PAGE), offer potential for improving health traits by targeting specific quantitative trait nucleotides (QTN) (Jenko et al., 2015).
4.3 Addressing reproductive efficiency
Reproductive efficiency is another area where quantitative genetics is applied in livestock breeding. Traits related to reproduction, such as fertility and litter size, are complex and influenced by many genetic and environmental factors. By utilizing quantitative genetic principles, breeders can identify genetic markers associated with reproductive traits and select animals with superior breeding values (Viana et al., 2016; Schmid and Bennewitz, 2017). This approach helps in optimizing reproductive performance and increasing the overall productivity of livestock populations. Techniques like genomic selection, which uses SNP markers to predict breeding values, have shown promise in improving reproductive traits.
In summary, quantitative genetics provides a framework for enhancing productivity, health, and reproductive efficiency in livestock breeding. By leveraging genetic information and advanced breeding techniques, breeders can achieve significant improvements in these key areas, ultimately leading to more efficient and sustainable livestock production systems.
5 Case Study: Application of Quantitative Genetics in Dairy Cattle Breeding
5.1 Background and significance of dairy cattle breeding
Dairy cattle breeding has evolved significantly over the past century, driven by the need to improve milk production and adapt to changing environmental and market demands. Initially, the focus was on increasing milk yield, but over time, the breeding goals have expanded to include traits such as fertility, health, and longevity, reflecting a more balanced approach to breeding (Figure 2) (Miglior et al., 2017; Weigel et al., 2017). The advent of genomic technologies has revolutionized the field, allowing for more precise selection and faster genetic progress (Gutierrez-Reinoso et al., 2021; Ma and Lin, 2024). This shift is crucial for meeting the growing global demand for dairy products while ensuring the sustainability and welfare of dairy cattle.
 Figure 2 Recording of performance data for dairy cows then (1936, left panels) and now (2017, right panel) (Adopted from Weigel et al., 2017) |
5.2 Use of heritability and breeding values in selecting high-yielding cows
Heritability and breeding values are fundamental concepts in quantitative genetics, used extensively in dairy cattle breeding to select high-yielding cows. Heritability estimates the proportion of phenotypic variation in a trait that is attributable to genetic differences among individuals. Traits such as milk yield have moderate to high heritability, making them suitable targets for selection (Kadri et al., 2015; Brajković et al., 2024). Breeding values, which predict an animal's genetic potential, are calculated using pedigree and performance data, and more recently, genomic information (Weigel et al., 2017; Gutierrez-Reinoso et al., 2021; Gutierrez-Reinoso et al., 2021). The integration of genomic selection has enhanced the accuracy of breeding values, enabling the identification of superior animals for milk production and other economically important traits.
5.3 Economic and practical impacts of quantitative genetics in the dairy industry
The application of quantitative genetics in dairy cattle breeding has had profound economic and practical impacts. By improving traits such as milk yield, health, and fertility, breeders have increased the productivity and profitability of dairy operations (Miglior et al., 2017; Weigel et al., 2017). Genomic selection has accelerated genetic gains, reduced generation intervals, and allowed for more efficient use of resources (Gutierrez-Reinoso et al., 2021). Additionally, the ability to select for disease resistance and adaptability to environmental stressors has improved animal welfare and reduced veterinary costs (König and May, 2019; Narayana et al., 2022). These advancements contribute to the sustainability of the dairy industry, ensuring it can meet future challenges and consumer demands.
In summary, the application of quantitative genetics in dairy cattle breeding has transformed the industry, enhancing productivity, sustainability, and animal welfare through the use of advanced genetic selection techniques.
6 Advances and Emerging Trends in Quantitative Genetics
6.1 Integration of genomics and big data in breeding programs
The integration of genomics and big data has revolutionized livestock breeding programs by enhancing the accuracy and efficiency of genetic selection. Genomic selection, which utilizes DNA sequencing and statistical algorithms, allows for the prediction of genomic breeding values, thereby accelerating genetic progress in complex traits such as growth and reproduction (Adebayo et al., 2024). The use of high-density SNP chips enables the selection of genetically superior animals at an early age with high accuracy, facilitating more precise breeding decisions (Singh et al., 2019). Additionally, the combination of genomic data with on-farm sensor data supports precision management, optimizing productivity and sustainability in modern dairy farms (Weigel et al., 2017).
6.2 Role of artificial intelligence and machine learning
Artificial intelligence (AI) and machine learning (ML) are playing increasingly significant roles in livestock breeding by improving the analysis and interpretation of complex genetic data. These technologies have been integrated into genomic selection processes, enhancing the prediction accuracy of breeding values and enabling more informed selection decisions (Weigel et al., 2017). Machine learning algorithms, alongside traditional mixed linear models, are now part of the modern breeder's toolkit, offering new insights into genetic inheritance and epigenetic modifications. The application of AI and ML in breeding programs is expected to continue growing, providing innovative solutions to challenges in genetic selection and management (Adebayo et al., 2024).
6.3 CRISPR and gene editing for precision breeding
CRISPR and other gene-editing technologies are emerging as powerful tools for precision breeding in livestock. These technologies allow for targeted modifications of specific genes, offering the potential to enhance desirable traits and eliminate undesirable ones with unprecedented precision. Gene editing can be used to knock out or over-express genes, providing deeper insights into gene functions and their impact on animal physiology (Khare and Khare, 2017). The application of CRISPR in livestock breeding holds promise for improving disease resistance, productivity, and overall animal welfare, marking a significant advancement in the field of quantitative genetics (Gutierrez-Reinoso et al., 2021).
In summary, the integration of genomics and big data, the application of AI and ML, and the use of CRISPR for gene editing are key emerging trends in quantitative genetics. These advancements are transforming livestock breeding by enhancing genetic selection accuracy, enabling precision management, and offering new possibilities for genetic improvement.
7 Challenges and Limitations
7.1 Complexity of traits and environmental interactions
Quantitative traits in livestock are influenced by numerous genes and environmental factors, making their genetic mapping and improvement complex. The interaction between these genes and the environment can significantly affect the expression of traits, leading to challenges in predicting breeding outcomes accurately. Genome-wide association studies (GWAS) have been employed to map these traits, but the nonadditive genetic and genotype-by-environment effects add layers of complexity that are difficult to unravel (Núñez-Torres and Almeida-Secaira, 2022). Additionally, the heritability of traits, which is crucial for effective breeding, can be obscured by these interactions, complicating the development of breeding strategies (Núñez-Torres and Almeida-Secaira, 2022).
7.2 Ethical and regulatory issues in advanced breeding technologies
The use of advanced molecular techniques, such as genome editing, raises ethical and regulatory concerns. While these technologies hold promise for improving quantitative traits, their application is often met with scrutiny regarding animal welfare and the potential long-term impacts on genetic diversity. The promotion of alleles through genome editing, for instance, can lead to increased inbreeding if not managed carefully, posing ethical dilemmas about the balance between genetic improvement and maintaining genetic diversity (Jenko et al., 2015). Regulatory frameworks are still evolving to address these issues, which can hinder the widespread adoption of such technologies (Kandel et al., 2023).
7.3 Accessibility and cost of genetic tools
The accessibility and cost of genetic tools remain significant barriers, particularly in developing regions. While molecular breeding techniques offer substantial potential for genetic improvement, their implementation is often limited by the high costs associated with genotyping and the need for sophisticated infrastructure (Kandel et al., 2023). This disparity in access can lead to unequal advancements in livestock breeding across different regions, with underdeveloped areas lagging behind due to financial and technological constraints (Khare and Khare, 2017). Moreover, the cost of routinely recording phenotypes and the sacrifice of animals for certain traits further limit the progress achievable through conventional breeding methods (Su et al., 2018).
In summary, the challenges in quantitative genetics for livestock breeding include the complexity of trait interactions, ethical and regulatory concerns surrounding advanced technologies, and the accessibility and cost of genetic tools. These factors collectively impact the efficiency and equity of breeding programs globally.
8 Future Directions
8.1 Expanding research on climate-resilient livestock breeds
The increasing impact of climate change on livestock production necessitates the development of climate-resilient breeds. Genomic tools and statistical models, such as genome-wide association studies (GWAS), have been instrumental in identifying traits that contribute to climate resilience in dairy cattle, including heat tolerance and disease resistance (Silpa et al., 2021; Arya et al., 2024). Additionally, integrating enviromics and machine learning can enhance precision breeding by considering the complex interactions between genetics and environmental factors, thus improving the adaptability of beef cattle to diverse climates (Passamonti et al., 2021; Rosa et al., 2023). These approaches are crucial for sustaining livestock productivity in the face of changing environmental conditions.
8.2 Leveraging multi-omics approaches for improved genetic predictions
Multi-omics approaches, which include genomics, transcriptomics, proteomics, and metabolomics, offer comprehensive insights into the genetic architecture of economically important traits in livestock. By integrating these diverse data types, researchers can improve the accuracy of genomic predictions for complex traits such as feed efficiency and meat quality (An et al., 2017; Diniz and Ward, 2021). The Functional Annotation of Animal Genomes (FAANG) project exemplifies efforts to enhance the functional understanding of livestock genomes, thereby facilitating more precise breeding strategies (Verardo et al., 2023). Continued development of statistical models that incorporate multi-omics data will be essential for advancing genetic predictions and improving livestock production efficiency.
8.3 Enhancing collaboration between academia and industry
Strengthening collaboration between academic institutions and the livestock industry is vital for translating research findings into practical breeding programs. Open-source breeding approaches, where public and private entities share data and align their activities, can enhance the efficiency of breeding pipelines and accelerate genetic gains (Covarrubias-Pazaran et al., 2021). Such collaborations can also foster the implementation of innovative breeding strategies, such as genomic selection and precision agriculture, to address challenges like climate change and disease resilience (An et al., 2017; Knap and Doeschl-Wilson, 2020). By leveraging shared resources and expertise, academia and industry can jointly develop solutions that meet the evolving demands of livestock production.
In summary, future research in livestock breeding should focus on developing climate-resilient breeds, utilizing multi-omics approaches for better genetic predictions, and fostering collaboration between academia and industry. These strategies will be crucial for enhancing the sustainability and productivity of livestock systems in the face of global challenges.
9 Concluding Remarks
Quantitative genetics plays a crucial role in livestock breeding by focusing on economically important traits such as body weight gain, milk, and meat production, which are characterized by continuous variability and are influenced by multiple genes and environmental factors. The application of genome-wide association studies (GWAS) and genome editing (GE) has advanced the understanding and improvement of these traits by identifying specific genes and mutations that contribute to genetic variance. Heritability remains a key factor in optimizing genetic quality, and various breeding methods, including panmixia, inbreeding, and heterosis, are employed to enhance selection processes. Additionally, the integration of molecular techniques, such as marker-assisted selection (MAS) and genomic selection (GS), has improved the efficiency of breeding programs by enabling direct selection on genes or genomic regions.
Quantitative genetics is fundamental to sustainable livestock breeding as it provides the framework for understanding and manipulating the genetic architecture of complex traits. By leveraging quantitative genetic principles, breeders can enhance traits that are vital for economic productivity and sustainability, such as disease resistance and production efficiency. The use of quantitative genetics in conjunction with modern genomic tools allows for more precise selection and breeding strategies, which can lead to significant improvements in livestock performance and resilience. This approach not only supports the economic viability of livestock production but also contributes to environmental sustainability by optimizing resource use and reducing the ecological footprint of agricultural systems.
Continued research and innovation in quantitative genetics are essential to address the evolving challenges in livestock breeding. There is a need for further exploration of genome editing techniques and their potential to enhance quantitative traits while managing inbreeding. Additionally, the development of more sophisticated statistical models and experimental designs for GWAS will improve the detection of nonadditive genetic effects and genotype-by-environment interactions. Collaborative efforts and open-source data management can accelerate genetic gains by facilitating the sharing of knowledge and resources among breeding programs. As the demand for food increases and climate change impacts production conditions, it is imperative to strengthen breeding pipelines with a focus on quantitative genetics to ensure the sustainability and resilience of livestock systems.
Acknowledgements
Author would like to express our gratitude to the two anonymous peer reviewers for their critical assessment and constructive suggestions on our manuscript.
Conflict of Interest Disclosure
Author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Adebayo O., Popoola M., Kuusu D., Fanwo R., Shoyombo A., Ndiomu E., Egbeyan J., and Moses A., 2024, Application of bioinformatics in animal breeding and genetics: a review, 2024 International Conference on Science, Engineering and Business for Driving Sustainable Development Goals (SEB4SDG), pp.1-7.
https://doi.org/10.1109/SEB4SDG60871.2024.10629845
An N., Lee S., Park J., Chai H., Cho Y., and Lim D., 2017, Current status of genomic prediction using Multi-omics data in livestock, Journal of Biomedical and Translational Research, 18(1): 151-156.
https://doi.org/10.12729/JBTR.2017.18.4.151
Arya A., Sharma P., Trivedi M., Modi R., and Patel Y., 2024, A look at genomic selection techniques for climate change adaptation and production in livestock, Journal of Scientific Research and Reports, 123(3): 307-317.
https://doi.org/10.9734/jsrr/2024/v30i62059
PMID: 30886391 PMCID: PMC6781144
Bijma P., 2021, 58 Integrating quantitative genetics and epidemiology: why selection against infectious diseases is more promising than we think, Journal of Animal Science, 99(Suppl 3): 31.
https://doi.org/10.1093/jas/skab235.054
Brajković V., Pocrnić I., Kapš M., Špehar M., Cubric-Curik V., Ristov, S., Novosel D., Gorjanc G., and Curik I., 2024, Quantifying the effects of the mitochondrial genome on milk production traits in dairy cows: empirical results and modelling challenges, Journal of Dairy Science, 108(1): 664-678.
https://doi.org/10.3168/jds.2024-25203
Covarrubias-Pazaran G., Martini J.W.R., Quinn M., and Atlin G., 2021, Strengthening public breeding pipelines by emphasizing quantitative genetics principles and open source data management, Frontiers in Plant Science, 12: 681624.
https://doi.org/10.3389/fpls.2021.681624
PMID: 34326855 PMCID: PMC8313805
Diniz W., and Ward A., 2021, 282 Multi-omics approaches to improve animal production, Journal of Animal Science, 99: 20-21.
https://doi.org/10.1093/JAS/SKAB054.036
Hill W., 2010, Understanding and using quantitative genetic variation, Philosophical Transactions of the Royal Society B: Biological Sciences, 365(1537): 73-85.
https://doi.org/10.1098/rstb.2009.0203
PMID: 20008387 PMCID: PMC2842708
Hayes B., and Goddard M.E., 2001, The distribution of the effects of genes affecting quantitative traits in livestock, Genetics Selection, Evolution, 33(3): 209-229.
https://doi.org/10.1186/1297-9686-33-2-191
PMID: 11403745 PMCID: PMC2705405
Gutierrez-Reinoso M.A., Aponte P.M., and García-Herreros M., 2021, Genomic analysis, progress and future perspectives in dairy cattle selection: a review, Animals, 11(3): 599.
https://doi.org/10.3390/ani11030599
PMID: 33668747 PMCID: PMC7996307
Haley C., 2002, Quantitative trait loci analysis in animals, Heredity, 88: 486-486.
https://doi.org/10.1038/sj.hdy.6800068
Halladakeri P., Gudi S., Akhtar S., Singh G., Saini D., Hilli H.J., Sakure A., Macwana S., and Mir R., 2023, Meta‐analysis of the quantitative trait loci associated with agronomic traits, fertility restoration, disease resistance, and seed quality traits in pigeonpea (Cajanus cajan L.), The Plant Genome, 16(3): e20342.
https://doi.org/10.1002/tpg2.20342
Jenko J., Gorjanc G., Cleveland M., Varshney R., Whitelaw C., Woolliams J.A., and Hickey J.M., 2015, Erratum to: potential of promotion of alleles by genome editing to improve quantitative traits in livestock breeding programs, Genetics, Selection, Evolution, 47(1): 69.
https://doi.org/10.1186/s12711-015-0135-3
PMID: 26361936 PMCID: PMC4567789
Kadri N., Guldbrandtsen B., Lund M., and Sahana G., 2015, Genetic dissection of milk yield traits and mastitis resistance quantitative trait loci on chromosome 20 in dairy cattle, Journal of Dairy Science, 98(12): 9015-9025.
https://doi.org/10.3168/jds.2015-9599
Kandel R., Kadariya I., Bohara K., and Adhikari S., 2023, A review on molecular breeding techniques: crucial approach in livestock improvement, Archives of Agriculture and Environmental Science, 8(4): 639-651.
https://doi.org/10.26832/24566632.2023.0804027
Khalil M., 2020, Molecular applications of candidate genes in genetic improvement programs in livestock, Egyptian Journal of Animal Production, 57(Suppl. Issue): 1-23.
https://doi.org/10.21608/ejap.2020.97954
Khare V., and Khare A., 2017, Modern approach in animal breeding by use of advanced molecular genetic techniques, International Journal of Livestock Research, 7(5): 1-22.
https://doi.org/10.5455/IJLR.20170404010154
Khatib H., and Gonda M., 2015, Molecular and quantitative animal genetics, John Wiley & Sons, pp.2-20.
Knap P.W., and Doeschl-Wilson A., 2020, Why breed disease-resilient livestock, and how? Genetics, Selection, Evolution, 52(1): 60.
https://doi.org/10.1186/s12711-020-00580-4
PMID: 33054713 PMCID: PMC7557066
König S., and May K., 2019, Invited review: phenotyping strategies and quantitative-genetic background of resistance, tolerance and resilience associated traits in dairy cattle, Animal, 13(5): 897-908.
https://doi.org/10.1017/S1751731118003208
PMID: 30523776
Li B., Vanraden P.M., Null D.J., O’Connell J.R., and Cole J., 2020, Major quantitative trait loci influencing milk production and conformation traits in Guernsey dairy cattle detected on BTA19, Journal of Dairy Science, 104(1):550-560.
https://doi.org/10.3168/jds.2020-18766
PMID: 33189290
Ma L., Cole J.B., Da Y.A.N.G., and VanRaden P.M., 2019, Symposium review: Genetics, genome-wide association study, and genetic improvement of dairy fertility traits, Journal of Dairy Science, 102(4): 3735-3743.
https://doi.org/10.3168/jds.2018-15269
Ma Q.X., and Lin X.F., 2024, From GWAS to breeding practice: genetic research on improving milk production in cattle, Animal Molecular Breeding, 14(1): 27-35
https://doi.org/amb.2024.14.0004
Miglior F., Fleming A., Malchiodi F., Brito L., Martin P., and Baes C., 2017, A 100-year review: Identification and genetic selection of economically important traits in dairy cattle, Journal of Dairy Science, 100(12): 10251-10271.
https://doi.org/10.3168/jds.2017-12968
Narayana S.G., de Jong E., Schenkel F.S., Fonseca P.A.S., Chud T., Powel D., Wachoski-Dark G., Ronksley P.E., Miglior F., Orsel K., and Barkema H., 2022, Underlying genetic architecture of resistance to mastitis in dairy cattle: a systematic review and gene prioritization analysis of genome-wide association studies, Journal of Dairy Science, 106(1): 323-351.
https://doi.org/10.3168/jds.2022-21923
PMID: 36333139
Núñez-Torres O., and Almeida-Secaira R., 2022, Quantitative genetics: principles of farming in livestock production, Journal of the Selva Andina Animal Science, 9(1): 23-36.
https://doi.org/10.36610/j.jsaas.2022.090100023x
Passamonti M.M., Somenzi E., Barbato M., Chillemi G., Colli L., Joost S., Milanesi M., Negrini R., Santini M., Vajana E., Williams J., and Ajmone-Marsan P., 2021, The quest for genes involved in adaptation to climate change in ruminant livestock, Animals, 11(10): 2833.
https://doi.org/10.3390/ani11102833
PMID: 34679854 PMCID: PMC8532622
Rosa G., Lourenco D., Rowan T., Brito L., Gondro C., Huang J., and De Souza S., 2023, 68 Integrating enviromics, genomics, and machine learning for precision breeding of resilient beef cattle, Journal of Animal Science, 101(Suppl 3): 49.
https://doi.org/10.1093/jas/skad281.060
Rukundo P., Karangwa P., and Uzayisenga B., 2018, Is quantitative genetics still necessary in this age of genomics? African Journal of Agricultural Research, 13(24):1227-1232.
https://doi.org/10.5897/ajar2017.12746
Schmid M., and Bennewitz J., 2017, Invited review: genome-wide association analysis for quantitative traits in livestock-a selective review of statistical models and experimental designs, Archives Animal Breeding, 60: 335-346.
https://doi.org/10.5194/AAB-60-335-2017
Silpa M.V., König S., Sejian V., Malik P., Nair M., Fonsêca V., Maia A., and Bhatta R., 2021, Climate-resilient dairy cattle production: applications of genomic tools and statistical models, Frontiers in Veterinary Science, 8: 625189.
https://doi.org/10.3389/fvets.2021.625189
da Silva F.L., Resende M.D.V., Ludke W.H., and Bueno T., 2017, Quantitative traits in breeding, Soybean Breeding, 2017: 81-112.
https://doi.org/10.1007/978-3-319-57433-2_6
Singh B.D., and Singh A.K., 2015, Mapping of quantitative trait loci, Marker-Assisted Plant Breeding: Principles and Practices, 2015: 185-216.
https://doi.org/10.1007/978-81-322-2316-0_7
Singh B., Mal G., Gautam S., and Mukesh M., 2019, Whole-genome selection in livestock, Advances in Animal Biotechnology, 2019: 349-364.
https://doi.org/10.1007/978-3-030-21309-1_31
Su H., Bijma P., Werf J., and Dekkers J.C.M., 2018, Software development for deterministic prediction of selection response in livestock breeding programs using genomic information, Journal of Animal Science, 96: 19.
https://doi.org/10.1093/JAS/SKY073.033
Van Den Berg I., Boichard D., and Lund M.S., 2016, Comparing power and precision of within-breed and multibreed genome-wide association studies of production traits using whole-genome sequence data for 5 French and Danish dairy cattle breeds, Journal of Dairy Science, 99(11): 8932-8945.
https://doi.org/10.3168/jds.2016-11073
PMID: 27568046
Verardo L.L., Brito L.F., Carolino N., and Magalhães A.F.B., 2023, Editorial: omics applied to livestock genetics, Frontiers in Genetics, 14: 1155611.
https://doi.org/10.3389/fgene.2023.1155611
PMID: 36873944 PMCID: PMC9978907
Viana J.M.S., Piepho H.P., and Silva F.F., 2016, Quantitative genetics theory for genomic selection and efficiency of breeding value prediction in open-pollinated populations, Scientia Agricola, 73(3): 243-251.
https://doi.org/10.1590/0103-9016-2014-0383
Weigel K.A., Vanraden P.M., Norman H.D., and Grosu H., 2017, A 100-year review: Methods and impact of genetic selection in dairy cattle-From daughter-dam comparisons to deep learning algorithms, Journal of Dairy Science, 100(12): 10234-10250.
https://doi.org/10.3168/jds.2017-12954
PMID: 29153163
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Jue Huang
Related articles
. Quantitative genetics
. Livestock breeding
. QTL mapping
. Genome-wide association studies
. Heritability
Tools
. Post a comment


